How To Filter Duplicate Contig
- Enquiry
- Open up Access
- Published:
Removing duplicate reads using graphics processing units
BMC Bioinformatics volume 17, Article number:346 (2016) Cite this article
Abstract
Background
During library structure polymerase chain reaction is used to enrich the Dna earlier sequencing. Typically, this process generates indistinguishable read sequences. Removal of these artifacts is mandatory, as they can affect the right interpretation of information in several analyses. Ideally, duplicate reads should exist characterized by identical nucleotide sequences. All the same, due to sequencing errors, duplicates may too exist nearly-identical. Removing about-identical duplicates can result in a notable computational effort. To deal with this challenge, we recently proposed a GPU method aimed at removing identical and virtually-identical duplicates generated with an Illumina platform.
The method implements an approach based on prefix-suffix comparing. Read sequences with identical prefix are considered potential duplicates. Then, their suffixes are compared to identify and remove those that are actually duplicated.
Although the method tin can be efficiently used to remove duplicates, there are some limitations that need to be overcome. In particular, it cannot to detect potential duplicates in the result that prefixes are longer than 27 bases, and information technology does not provide support for paired-cease read libraries. Moreover, large clusters of potential duplicates are split into smaller with the aim to guarantees a reasonable computing time. This heuristic may bear on the accuracy of the assay.
Results
In this work we propose GPU-DupRemoval, a new implementation of our method able to (i) cluster reads without constraints on the maximum length of the prefixes, (ii) support both single- and paired-end read libraries, and (iii) analyze large clusters of potential duplicates.
Conclusions
Due to the massive parallelization obtained by exploiting graphics cards, GPU-DupRemoval removes duplicate reads faster than other cutting-edge solutions, while outperforming most of them in terms of amount of duplicates reads.
Groundwork
Duplicate reads are one of the most problematic artifacts generated during polymerase chain reaction distension. Ideally, duplicates should take identical nucleotide sequences. However, due to sequencing errors, they may end upwards to exist nearly-identical [1]. Duplicates can bear on the accuracy of some analyses on NGS data. Removal of these artifacts tin be an essential pre-processing step, in detail on applications based on resequencing. For instance, in SNP calling, errors introduced in early amplification steps are shared by PCR duplicates, making very difficult to distinguish between repeated (but identical) errors and existent SNPs [ii, 3]. Duplicate removal is also a mandatory stride to find CNVs using read-depth (RD) based methods [four]. These methods assume that the RD in a genomic region depends on the re-create number of that region. As a consequence, duplicates demand to be detected and removed to avert incorrect read count. Duplicates can as well affect the accuracy of de-novo sequencing. During scaffolding, paired-finish reads are mapped on contigs with the aim to rank their guild. In this stage, 2 contigs are considered connected depending on the number of read pairs that link them (the higher the number the stronger the connection). Hence, PCR duplicates may result in false-positive connections betwixt contigs.
Duplicate sequences can be natural or artificial. Ideally, only artificial duplicates should be removed, while natural ones should be retained. Unfortunately, natural and artificial duplicate sequences are indistinguishable. This is the reason why a fraction of reads labeled as duplicates may in fact be generated from distinct molecules, yielding a loss of natural reads. However, this situation occurs typically during the analysis of single-cease reads. In fact, as for paired-cease reads, the probability of finding contained molecules identical at both ends being very low [5].
Removal tools proposed in the literature implement methods that focus either on alignment-based or on alignment-free strategies. Alignment-based tools presume that duplicate reads will be mapped to the same position on a reference genome. These tools analyze the alignments obtained past running an embedded process (or a 3rd-party aligner) with the goal of finding reads with identical mapping coordinates. These reads are analyzed and those that meet predefined quality constraints are classified as duplicates. The performance of these tools is affected by the alignment constraints and by the accuracy of the aligner. Moreover, information technology should be pointed out that these tools cannot exist used in absence of a complete reference genome.
Picard MarkDuplicates [6], samtools rmdup [7], and SEAL [8] are tools that implement an alignment-based strategy. Picard MarkDuplicates identifies duplicates by analyzing the alignments generated by a third-political party aligner. Every bit for paired-stop reads, it finds the 5' coordinates and mapping orientations of each read pair. All pairs with identical coordinates and orientations are analyzed and those having the highest sum of base of operations qualities are classified equally duplicates. Information technology as well removes duplicates from single-finish libraries. Similarly, the rmdup function of samtools analyzes alignments obtained with a third-party tool to remove duplicates from both single- and paired-end reads. However, differently from Picard MarkDuplicates, rmdup is not able to remove interchromosomal duplicate reads. SEAL provides a distributed version of BWA [9] to perform the alignment and removes duplicates according to the same criteria employed by Picard MarkDuplicates.
Alignment-free tools detect duplicates by comparing read sequences. In item, those reads characterized by a similarity score higher than a given threshold are classified as duplicates. Notably, tools that comply with this strategy are not affected by the bias introduced past a brusque-read mapping tool and can besides be used in absenteeism of a complete reference genome. Unfortunately, they may be computationally onerous, as each sequence of the dataset must be compared to all other sequences in the dataset. This is the reason why heuristics are defined and adopted to deal with the computational challenge.
Fastx-Toolkit Collapser [10], FastUniq [11], Fulcrum [12] and CD-HIT [13–15] are all examples of tools that implement an alignment-free strategy. Fastx-Toolkit Collapser is able to identify and remove identical sequences from single-terminate reads. Conversely, FastUniq has been designed to remove identical duplicates from paired-end reads. Removal is performed executing three steps in pipeline. Initially, all paired reads are loaded into retention. Then, read pairs are sorted according to their nucleotide sequences. Finally, duplicates are identified by comparing the adjacent read pairs in the sorted list. Fulcrum is able to place identical and virtually-identical duplicates from both single- and paired-end reads. It identifies as potential duplicates those reads with an identical prefix of the nucleotide sequences. Potential indistinguishable reads are binned in unlike files, whose maximum size is user-divers. Read sequences inside each file are compared to identify duplicates. CD-HIT provides two unlike tools to remove duplicates from single- and paired-cease reads generated with 454 or Illumina platform. CD-HIT-454 analyzes libraries generated with 454 to place duplicates that are either exactly identical or come across the following criteria: a) reads must be aligned at 5'-ends; b) for sequences of dissimilar length, a shorter read must exist fully aligned with the longer one (the seed) and they accept less than user-defined percentage of indels and substitutions. CD-Hitting-DUP removes duplicates from Illumina libraries analyzing the prefix of the read sequences. Read sequences with identical prefix are considered duplicated. For paired-finish reads, prefixes at both ends are checked. Features of the listed tools are summarized in Table 1.
Recently, nosotros proposed a new alignment-free method aimed at removing duplicate reads using Graphics Processing Units (GPUs) [16] generated with an Illumina platform. In particular, nosotros implemented a prefix-suffix comparison algorithm which takes into business relationship the per-base error rates generate with Illumina. The method consists of two phases, which accept been massively parallelized on GPU. Initially, potential duplicate sequences are amassed according to their prefix. And so, the suffixes of the sequences in each cluster are compared to detect and remove duplicates.
Although the method can be efficiently used to remove both identical and nearly-identical duplicates, there are some constraints and limitations that need to be overcome. In detail, information technology does not allow to detect potential duplicates on prefixes longer than 27 bases, does not support paired-stop read libraries, and imposes a constraint on the maximum size of the clusters.
In this work we nowadays GPU-DupRemoval (standing for GPU-Duplicates Removal) a new implementation of our method devised to overcome these limitations. In particular, i) cluster reads without constraints on the maximum length of the prefixes are now allowed, ii) back up for both single- and paired-cease read libraries is provided, and three) larger clusters of potential duplicates (without using heuristics) can now be candy.
Implementation
Before going into relevant details of the proposed algorithm, let us requite a brusk introduction to GPUs.
Graphics processing units
GPUs are hardware accelerators that are increasingly used to deal with different computationally intensive bioinformatics algorithms (e.g., [17–21]). From an architectural perspective, the main departure between traditional CPUs and GPUs is related to the number of available cores. Indeed, the former are devices composed of few cores, with lots of cache retention able to handle a few software threads at a time. Conversely, the latter are devices equipped with hundreds of cores able to handle thousands of threads simultaneously, so that a very high level of parallelism can be reached.
The intensive use of GPUs over the concluding years has yielded a significant increases in the performance of several applications. However, it should be noted that but algorithms based on the SIMD paradigm tin can be effectively parallelized on GPUs. CPUs and GPUs should be considered equally complementary for dissimilar types of processing. CPUs are optimized for period control and low memory latency, whereas GPUs are optimized for information parallel computations. In this context, the GPU computing model uses CPUs and GPUs in a heterogeneous co-processing calculating model. Computationally-intensive parts of an algorithm based on the SIMD image tin be accelerated by GPUs, whereas CPU is used to control the GPU execution while processing other parts of the algorithms not suitable for the GPU.
Equally for GPU programming, CUDA (Compute Unified Device Architecture) [22] and OpenCL (Open Computing Linguistic communication) [23] offer ii unlike interfaces for GPU programming. It is worth pointing out that OpenCL is an open standard that can be used to programme CPUs, GPUs and other devices from dissimilar vendors whereas CUDA is specific to NVIDIA GPUs.
The algorithm
Assay of short-read datasets generated with Illumina highlighted a very low charge per unit of indel errors (<0.01 %) while the number of occurrences of wrong bases increases with the base position [24]. Therefore, it is possible to deduce that: (i) the bulk of duplicates will differ on few base of operations substitutions; (ii) most of identical and nearly-identical duplicates are in fact characterized by an identical prefix. Starting from these considerations, we devised a method aimed at comparison only potential indistinguishable reads (i.e., reads with identical prefix) without taking into account indels in sequence comparisons.
Initially, potentially duplicated sequences are clustered together (see Fig. one). And so, for each cluster, the first sequence is taken every bit a seed and its suffix is compared with those of the other sequences that fall in the selected cluster. Sequences that are identical or very similar to the seed are classified as duplicates. Duplicates are condensed in a new sequence and are removed from the cluster (come across Fig. two). Then, the process is iterated for the remaining sequences in the cluster (if any), until the cluster is empty or contains only one read sequence.
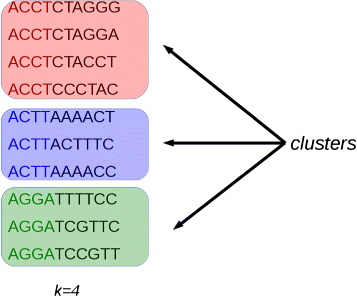
Clustering. Reads with an identical prefix of k nucleotides are considered potential indistinguishable reads. Image from [sixteen] used under the terms of the Creative Commons Attribution License (CC Past)
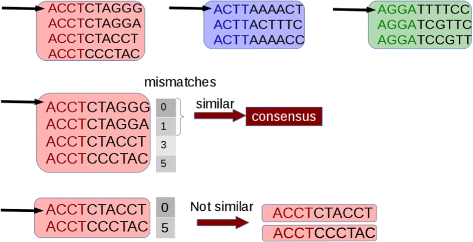
Comparing. The first read of each cluster is taken as a seed and its suffix is compared with that of the other sequences in the cluster. Sequences that differ from the seed for a number of mismatches lower than a user-defined threshold are considered duplicates of the seed. Each gear up of duplicates is removed from the cluster and are represented with a consensus sequence. The process is iterated until the cluster is empty. Epitome from [16] used nether the terms of the Creative Eatables Attribution License (CC BY)
Clustering is performed by sorting the prefixes of the read sequences with our GPU-based CUDA-Quicksort [25]. As CUDA-Quicksort sorts numerical values, information technology is necessary to encode the prefixes of the read sequences. To this stop, we devised the encoding with the aim to represent as many nucleotides every bit possible with a numeric value. In as doing, read sequence prefixes are subject to a dual numeric encoding. Initially, prefixes are represented with a base-five encoding past replacing each nucleotide with a numerical value ranging from 0 to 4 (i.e., A→0,C→ane,G→ii,T→3,N→4). Representing items with 64 bit unsigned long long int data type allows to encode and sort prefixes of up to 19 nucleotides. A longer prefix would exceed the limit for this blazon of data. Information technology is possible to overcome this constraint using a different numerical base for representing prefixes. In item, converting to base-ten the prefixes encoded using a base-5 encoding, information technology is possible to correspond a number consisting of upwards 27 digits with a 64 bit unsigned long long int (encounter Fig. 3).
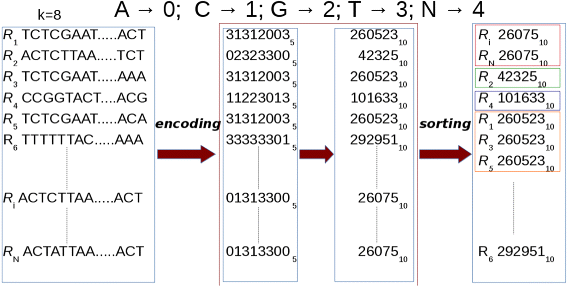
Enconding. Prefixes are subjected to a dual encoding. Initially, the nucleotides in a prefix are encoded with a numerical value from 0 to iv (i.east., A→0, C→1, G→2, T→3, N→iv). So, these numerical representations are encoded using base-x. Finally, sorting is performed for clustering. In the figure, prefixes of length one thousand=8 are represented. Paradigm from [sixteen] used under the terms of the Artistic Commons Attribution License (CC BY)
For the sake of completeness, information technology should be pointed out that regarding the problem addressed in this piece of work, quick-sort is more than effective than other sorting algorithms, including radix-sort. With k number of digits in a key and north number of keys, the computational complexity of comparison-based radix-sort is O(1000·northward), whereas the complexity of quick-sort is O(n·fifty o g(due north)). Hence, quick-sort outperforms radix-sort in the issue that thou>fifty o 1000(north), and viceversa. The performance of CUDA-Quicksort has been compared with Thrust Radix Sort [26], a cutting-edge algorithm running on GPUs. The comparative assessment has been made in the chore of sorting items with long keys -characterized by xix digits (i.due east., the maximum number of digits used to represent the encoded read prefixes). Experiments, performed ensuring a uniform distribution on benchmark datasets (with varying size from 1M to 32M elements), show that CUDA-Quicksort outperforms Thrust Radix Sort with a speed-upwards ranging from 1.58x to 2.18x, depending on the dataset at manus [25].
Despite the fact that longer sequences (i.east., 27 instead of 19 nucleotides) can be candy, the latter encoding is still restrictive. As the quality of Illumina reads decreases with the position that a base has in the sequence being processed, more than probable sequencing errors are localized towards the iii' cease of a read rather than in proximity of to the five' stop. In GPU-DupRemoval, two sequences are classified as nearly identical if they fulfill a given constraint on the maximum number of immune mismatches. In doing so, a mismatch is always considered a sequencing fault, irrespective of the position (and of the quality) that a base has in a read. This processing policy gives rise to a fraction of natural nearly-identical sequences that may be erroneously classified as artificial duplicates. An approach to reduce the number of false positives in the process of duplicate identification is to limit the analysis of mismatches where is more likely sequencing errors are localized, choosing the prefix length according to the resulting quality scores obtained across all bases on the dataset. This length must be chosen to permit the pick of all bases whose average quality score is higher than a given threshold.
Afterwards that reads have been clustered, their suffixes are compared. Basically, a base of operations-per-base comparison of the nucleotides of the suffix of the seed with those of the other reads in a cluster should exist performed in this phase. This approach might crave a very high number of comparisons. Permit Due north exist the length of the suffixes, and let yard be the minimum number of mismatches immune to consider two sequences as non duplicated. In the best example, two sequences can be classified as not duplicated after yard comparisons. In the worst case N comparisons must be performed. Nosotros implemented a different strategy aimed at reducing the number of comparisons. Initially, suffixes are dissever into fixed-length chunks. Each subsequence representative of a chunk is subjected to the same dual numeric encoding used to represent the prefixes for clustering. And then, for each cluster, the numerical difference between the i-thursday chunk of the seed and the related chunk of the other suffixes in a cluster is calculated (meet Fig. 4). The order of magnitude of this difference provides information about the position of the leftmost different nucleotides. And so, subsequences are cutting at the mismatch position. The rightmost parts of the mismatch position are maintained and the process is re-iterated. In the worst case, this approach is able to classify two sequences as not duplicated after m comparisons.
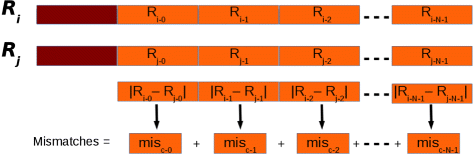
Suffixes. Suffixes (in orange in the figure) are analyzed in chunks. Each clamper is subject to the dual encoding used for prefixes (in red in the effigy). The overall number of mismatches if obtained summing the partial number of mismatches obtained for each clamper. Image from [16] used under the terms of the Creative Commons Attribution License (CC Past)
Suffix comparison has been massively parallelized on GPU. In particular, the chunks representatives of the reads in a cluster are loaded into the GPU retentiveness and compared in parallel with the chunk of the seed. It should be noted that also the size of the clusters affects the overall computing fourth dimension. In fact, depending on both the size of a cluster and the percentage of duplicates in information technology, a very high number of comparisons amid sequences could be performed. In our method, very large clusters are split into smaller ones of fixed size with the aim of reducing the number of comparisons. In a similar manner, in Fulcrum, potential duplicates are binned in file of user-defined maximum size. On one hand this heuristic guarantees a reasonable calculating time. However, on the other hand, information technology may bear upon the accuracy of the assay. Therefore, in our view, resorting to this heuristic appears not appropriate. In the following, we describe the changes implemented in GPU-DupRemoval to cope with the constraints on the prefix length and the maximum size of a cluster, and to support paired-end reads.
Prefix length
The proposed clustering strategy resulted be very effective. Its computing time depends on the size of the dataset, whereas information technology does not depend on the prefixes length. Moreover, CUDA-Quicksort is able to cluster datasets of millions of reads very apace. Information technology should be pointed out that CUDA-Quicksort resulted be the faster implementation of the sorting algorithm on GPUs. In particular, it was up to 3 times faster than the CDP-Quicksort released past NVIDIA.
Starting from these considerations, we devised a multi-stride clustering strategy based on the existing i. For prefix length of up to 27 nucleotides clustering does not differ from the previous version of the tool. The approach differs when longer prefixes must be analyzed. Initially, prefixes are carve up into chunks of 27 nucleotides. Obviously, depending on the length of the prefixes, the final chunk might be shorter than 27 nucleotides. Each prefix is subjected to the dual numerical encoding previously described. So, CUDA-Quicksort is used to sort reads according to two different criteria. The first sorting (say A) is obtained according to the kickoff chunk of prefixes. It represents the partial sorting of the reads that will be iteratively updated to build the last sorting. The second sorting (say B) is obtained according to the second chunk of the prefixes. This sorting is used to update the commencement 1 in such a way that it becomes a sorting representative of both chunks. Basically, B is used to update the ordering of the reads in the clusters generated according to A. To this stop, an array is initialized to store the new sorting (say C). The array will exist partitioned taking into account the clusters generated with A. And then, according to B, the reads of each cluster are copied in the new assortment. Each read is copied in the kickoff free position of the partition related to its belonging cluster generated with A (see Fig. 5). Later that all reads in a cluster generated with B have been copied into the new assortment, both the number of clusters and their size is updated. A cluster is carve up into two clusters each time that its related partition in the new array is just partially written. At the end of the procedure, A is replaced by C. Then, the process is re-iterated with the following chunks (if whatever).
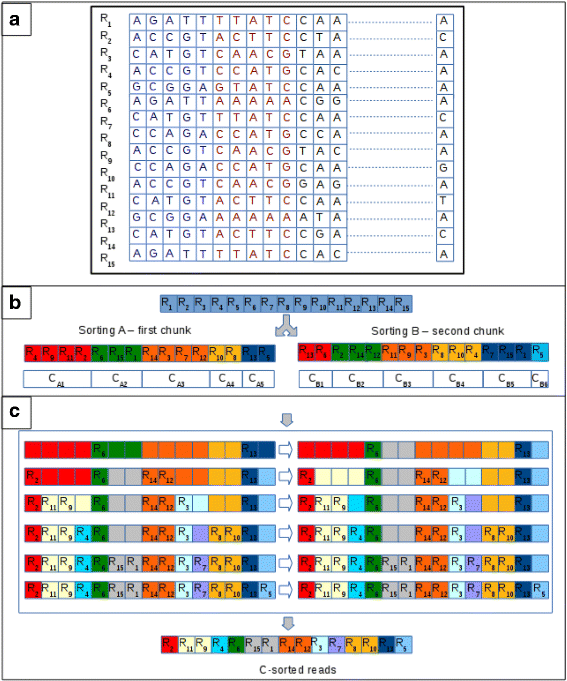
Multi-pace clustering. To simplify the graphical representation, we assume that the multi-stride clustering is enabled for prefixes longer than 5 nucleotides. In this example 15 reads are clustered analyzing prefixes of ten nucleotides. a Initially, the prefixes are carve up into two chunks of five nucleotides. In the effigy, the nucleotides of the first chunk are represented in blue, and those of the 2d chunk are represented in red. The clustering consists of three steps. b Reads are clustered past sorting them according to the first chunk of the prefixes (sorting A in the figure). Clustering generates 5 clusters of different size (C A1,C Atwo,C Aiii,C Aiv,C Afive). Reads clustered together are represented with the same background color. Afterward, reads are clustered by sorting them according to the second chunk of the prefixes (sorting B in the figure). This clustering generates 6 clusters unrelated from those of the previous clustering (C B1,C B2,C B3,C B4,C B5,C B6). c A new array is initialized and partitioned according to the size of the clusters of A. The sequences of each cluster in B are copied in the new array in the partition associated to their belonging cluster in A. Each read is copied into the first free position of the division. The process is represented in the C box. Each row reported therein represents the process of copying the reads of a cluster in C. On the left it is shown where the reads are copied, whereas on the right information technology is shown how clusters are divide later each iteration. Initially, the reads R 13 and R 6 of C B1 are copied in the new array. R thirteen belongs to C A5 in A and R half dozen belongs to C A2 in A. Existence the first reads to exist analyzed, they are copied in the kickoff position related to its cluster in A. Cluster C A2 and C A5 are partially filled later this step. This implies that the reads in these clusters are non identical, according to the 2nd chunk of their prefix. In fact, R 15 and R 1 have not been clustered together with R 6 in B. Similarly, R 6 has not been clustered together with R thirteen in B. Therefore, the clusters are carve up (see first row in C box). Cluster C A2 is split into two clusters. A cluster contains R 6 and the other cluster (of size 2) is empty. Similarly, C A5 is split into two clusters of size ane. The process is iterated (as represented in the C box) until all clusters in B accept been analyzed. The final sorting generates 11 clusters
Information technology might seem that the described algorithm implements a radix sort. However, in that location are considerably differences between the two algorithms. Radix-sort is a non-comparative sorting algorithm that performs a digit-by-digit sorting on keys. In the proposed multi-step clustering strategy, GPU-DupRemoval uses CUDA-Quicksort to perform (at each step) a comparing-based sorting of numbers (not digits), representative of sub-sequences of the available reads. Furthermore, let us recall that two variants of the radix-sort (RS) be, able to process keys from the less to the most pregnant digit (i.east., LSD/RS) and from the about to the less significant digit (i.due east., MSD/RS). The apparent resemblance of multi-step clustering algorithm with radix-sort in fact applies more to LSD/RS than to MSD/RS. Withal, but MSD/RS could be used to address the problem at hand as, unlike LSD/RS, MSD/RS dispatches all digits with identical value into a specific bucket and recursively repeats the same operation with all buckets, until sorting is complete. Notably, keys that occur in a saucepan are sorted independently from those in other buckets. Conversely, in the proposed approach, sorting is performed by analyzing all sequences at each step.
This multi-stride clustering strategy has been used to optimize besides the removal of identical duplicate reads. In the first implementation the same approach was used to remove identical and virtually-identical reads. However, identical reads tin can be removed more hands than virtually-identical ones. In particular, identical reads can be identified past clustering the reads by their entire sequences. In GPU-DupRemoval identical duplicates are automatically removed by clustering reads according to their entire sequences. It should be pointed-out that the multi-step clustering is non used to remove identical duplicates when GPU-DupRemoval is run to remove both identical and virtually-identical read sequences. It is solely used when GPU-DupRemoval is used to remove identical reads.
Maximum size of a cluster
Equally previously described, in the commencement implementation of our method large clusters are automatically dissever into smaller. On ane hand, analyzing pocket-size clusters may improve the operation in terms of computing time; on the other manus, it may worsen the performance in terms of accurateness. The smaller the cluster is, the faster the processing is, as fewer comparisons are required. Unfortunately, duplicates separated during the splitting will be not identified.
The problem has been addressed in GPU-DupRemoval, which is able to analyze large clusters without the demand for splitting them. Originally, only a level of parallelism was implemented. At each iteration, the first read in each cluster (i.e., the seed) is compared with the other reads in the cluster. Depending on both the size of a cluster and on the per centum of duplicates, this approach may require many iterations. Allow N be the size of the cluster, when each read is uniquely represented in the cluster (worst case), N-1 iterations must be performed. This computational challenge can exist efficiently addressed past calculation a second level of parallelism aimed at comparing multiple seeds of a cluster in a single iteration. In the current implementation, at each iteration the possibility to compare in parallel multiple seeds of a cluster is assessed. Information technology should be pointed out that depending on the size of the dataset at hand, an iteration might require one or more kernel launches. Notably, GPU-DupRemoval applies different strategies, depending on the number of kernel launches required to analyze the dataset. Initially, GPU-DupRemoval determines the thread block size, the number of thread blocks and the GPU retentivity required to clarify the given dataset and to compare the read chunks that occur in each cluster with a unmarried seed. Every bit long every bit more than ane kernel launch is required in an iteration, GPU-DupRemoval compares reads that occur in a cluster with a single seed. Conversely, when a single kernel launch is made in an iteration, GPU-DupRemoval checks the feasibility of comparison multiple seeds in parallel. The upper limit of seeds that can be compared in parallel for each cluster (say n) is determined according to the constraints on the maximum number of blocks that can exist created per kernel launch and on the memory of the device. At each iteration, up to north seeds are compared in parallel for each cluster. The upper limit for the seeds that can exist compared in parallel will increment with a subtract of the read sequences to be analyzed. Every bit the number of reads decreases at each iteration, the value of n is re-calculated afterward each kernel launch. In and so doing, when multiple seeds are analyzed in parallel, a read is compared with two or more than seeds in the same iteration, and, depending on the results of comparisons, it may exist classified as duplicated of two or more seeds. In these cases, the read will be considered as duplicated of the first seed.
Supporting paired-end reads
GPU-DupRemoval has been devised to support both unmarried- and paired-terminate reads. Information technology should be noted that duplicates from paired-finish reads can be removed similarly to that concerning unmarried-end reads. In fact, paired-finish reads with an identical prefix at both ends tin be considered equally potential duplicates. Hence, potential duplicates can be identified by clustering reads according to the prefixes that occur at both ends. To this finish, GPU-DupRemoval builds a new sequence representative of both reads for each pair in the dataset. Each sequence is build by merging separately prefixes and suffixes, as shown in Fig. 6. Afterwards, these sequences are analyzed co-ordinate to the same method used for single-end reads. Finally, after that duplicates have been removed, sequences are demerged (see Fig. 7). It should be pointed out, that this strategy to back up paired-end reads is well suited to the current implementation of the algorithm that gives a viable solution to the result concerning the maximum length of prefixes. The limitation on the length of the prefixes of the previous implementation would negatively affect the capability of removing duplicates from paired-end reads.

Merging paired-end reads. Paired-finish reads with identical prefix at both ends can be considered potential duplicates. The same clustering strategy used to place potential duplicates in single-end reads can also be used for paired-terminate reads. In this instance, paired-terminate reads need to be merged every bit represented in the effigy. A sequence representative of a pair is obtained by merging the prefixes and the suffixes of both forward and opposite read. With N the length of the read sequence and p length of the prefixes, the new sequence consists of 2·N nucleotides and is represented past a prefix of 2·p nucleotides

De-merging paired-terminate reads. After that duplicates have been removed, sequences are demerged to generate both forrad and contrary reads
Results and discussion
Experiments have been designed to assess the performances of GPU-DupRemoval to remove identical and most-identical duplicates from unmarried- and paired-end read libraries, with both synthetic and real life data. In this section, we start introduce experiments on synthetic information, which are mainly aimed at assessing the reliability of GPU-DupRemoval. Experimental results obtained on real data are reported subsequently.
Functioning evaluation on synthetic data
Synthetic libraries have been generated with the Sherman simulator [27]. It should be pointed out in advance that Sherman does not let to prepare the percent of duplicates in a library. Initially, nosotros used Sherman to generate a library consisting of 750 thousands of 100 bp unmarried-finish reads. Subsequently, constructed reads have been candy to generate 250 thousands of 100 bp duplicate reads. Duplicates consist of identical duplicates (100 thousands), duplicates generated by simulating a i % of sequencing error (100 thousands), and duplicates generated by simulating a sequencing error ranging between 2 and 3 % (50 thousands). Similarly, nosotros generated a library consisting of i millions of 100 bp paired-end reads. In this instance, the sequencing mistake has been uniformly imitation on both ends. As for both single- and paired-finish reads, duplicates have been generated past simulating sequencing errors using an error rate curve that follows an exponential disuse model, with the aim of mimicking existent data.
Equally for the single-end library, GPU-DupRemoval has been compared with Fastx-Toolkit Collapser, CD-Hit-DUP, and Fulcrum. Experiments accept been divers to assess the reliability of the tool to identify and remove duplicates according to the sequencing errors injected therein. Results reported in Table two show the percentage of reads removed from each tool when used to identify duplicates co-ordinate to a sequencing fault ranging from 0 to three %. Apart from CD-Hit-DUP, the other tools work properly and take been able to identify all duplicates. Similar behavior has been observed for paired-terminate reads (see Tabular array 3). In this instance, GPU-DupRemoval has been compared with FastUniq, CD-Hitting-DUP, and Fulcrum. It should be pointed out that both Fastx Toolkit Collapser and FastUniq does not support removal of nearly-identical reads.
Operation evaluation on real data
To assess the performance of GPU-DupRemoval on real data, we used information technology to remove duplicates of 2 libraries generated with the Illumina platform; i.e., library SRR921897 consisting of 50 millions of 100 bp unmarried-finish reads, and library SRR005718 consisting of 32 millions of 36 bp paired-terminate reads. Experiments have been carried-out to identify and remove identical and well-nigh-identical duplicates with up to i and 3 mismatches.
Experiments described hereinafter have been carried out on a 12 cores Intel Xeon CPU E5-2667 two.90 GHz with 128 GB of RAM. An NVIDIA (Kepler compages based) Tesla k20c card with 0.71 GHz clock rate and equipped with 4.8 GB of global memory has been used to execute GPU-DupRemoval.
Experiments have been designed with the goal of providing a rigorous comparison amid the tools. In this context, it should be pointed out that Fulcrum considers as aNy those bases with a quality score under a user defined-threshold. Being non supported by the other tools, this option has been disabled as it can affect the percentage of duplicates removed. Moreover, differently from the other tools, Fulcrum parallelizes the ciphering on multiple CPU cores. Therefore, to provide a rigorous comparison in terms of computing time with GPU-DupRemoval, Fulcrum has been run parallelized on all available CPU cores.
Identical parameters have been used to perform the clustering in both GPU-DupRemoval and Fulcrum. Clustering has been performed co-ordinate to unlike lengths of the prefixes with the aim to show how this parameter affects the removal of duplicates. Every bit for nearly-identical duplicates, clustering has been performed by analyzing prefixes of 25/35/45/55 bases for SRR921897 and prefixes of 10/15 bases for SRR005718. As for identical duplicates, GPU-DupRemoval automatically clusters the reads past their entire nucleotide sequences. The same constraint has been used in Fulcrum to remove identical duplicates.
Tabular array 4 summarize the results obtained by removing duplicates from the SRR921897 library in terms of removed reads, computing time, and peak of retentiveness required. GPU-DupRemoval, CD-HIT-DUP, and Fastx Toolkit Collapser removed the same percentage of identical read sequences (vii.4 %). A slightly lower amount (vii.3 %) of identical reads has been removed past Fulcrum. Every bit for nearly-identical reads, Fulcrum removed a slightly higher amount of sequences with respect to GPU-DupRemoval. In particular, clustering the reads with a prefix length of 25/35/45/55 bases GPU-DupRemoval removed 9.2/viii.9/8.7/8.4 % of nearly-identical sequences with up to 1 mismatch, whereas the percentage reached by Fulcrum was nine.8/9.half-dozen/9.4/nine.2 %. Similar results accept been obtained to remove virtually-identical sequences with up to 3 mismatches. In this case, clustering the reads with a prefix length of 25/35/45/55 bases GPU-DupRemoval removed 12.2/11.5/10.eight/x.0 % of sequences, whereas the percentage of sequences removed by Fulcrum was 13.1/12.3/11.7/10.9 %. As for CD-Hit-DUP, it has been able to remove 8.0/9.viii % of duplicates with up to one/3 mismatches.
We deem that this slight discrepancy on the pct of sequences removed by GPU-DupRemoval and Fulcrum depends on the different strategies implemented to compare the reads in a cluster. In Fulcrum, initially, a list of groups of strongly similar reads is initialized using the beginning read of each cluster (say r). Subsequently, each read in a cluster is compared to r and if considered like to r it is added to the group and a new consensus sequence r is calculated and used for the following comparing. Unlike, GPU-DupRemoval does not compare the reads in a cluster with a consensus sequences. In GPU-DupRemoval duplicates are detected by comparing each read in a cluster to all other sequences in the cluster.
Results prove how the prefix length used for clustering can bear upon the pct of removed reads. Analysis of results show that the percent of sequences classified as duplicates decreases with increasing the prefix length. For case, with a prefix of 35 bases the pct of nearly-identical sequences with up to 1/3 mismatches removed past GPU-DupRemoval decreased of 0.three/0.7 % with respect to the percentage obtained using a prefix of 25 bases. Similar results have been obtained by Fulcrum.
Operation in terms of computing fourth dimension shows that GPU-DupRemoval is the fastest tool. It resulted to be 3.0x faster than Fastx Toolkit Collapser in removing identical duplicates, whereas it resulted to be upwards to four.2x/ii.1x/v.6x faster than CD-HIT-DUP and upward to 13.2x/4.1x/five.2x faster than Fulcrum to remove duplicates with up to 0/one/three mismatches.
As for memory consumption, Fulcrum outperforms the other tools, the worst beingness CD-Striking-DUP, which requires a notable loftier amount of retention. Experiments show that the memory required increases with the allowed number of mismatches. In particular, CD-Hitting-DUP required 33.3 GB of retentiveness to remove identical duplicates and 49.2/53.five GB of retention to remove duplicates with up to i/3 mismatches. As for GPU-DupRemoval, the amount of memory required depends on both the size of the dataset and the type of duplicates removed, whereas it is unrelated from the number of differences allowed among duplicates. To perform the experiments, GPU-DupRemoval required 13.one GB to remove identical duplicates and 16.6/17.two/sixteen.5/17.5 GB to remove near-identical sequences clustering the reads with a prefix length of 25/35/45/55 bases.
Experiments take too been performed to compare the performance of the current implementation of GPU-DupRemoval with that obtained with the previous release. Experiments with the get-go release of the algorithm has been performed clustering the reads with a prefix length of 25 bases. Every bit for identical duplicates, both releases of the tool have been able to remove the aforementioned percentage of identical duplicates. However, due to the multi-clustering strategy the current release of the tool resulted to be 5.5x faster than the previous implementation. Every bit for virtually-identical reads both versions of the tool exhibit comparable functioning in terms of computing time and retentiveness consumption. However, the electric current version of the tool, which is able to analyze large clusters without heuristics, has been able to remove a slightly higher per centum of duplicates.
Like results are reported in Table 5 for SRR005718 library. Equally for identical duplicates, all tools removed the aforementioned amount of reads (2.9 %). GPU-DupRemoval and Fulcrum show virtually identical results. GPU-DupRemoval removed iii.6/3.5 % of duplicates with up to 1 mismatch clustering the reads with a prefix length of 10/15 bp, whereas Fulcrum removed always 3.6 % of duplicates. As for nearly-identical duplicates with upwards to 3 mismatches GPU-DupRemoval removed iv.0/3.nine % of duplicates clustering the reads with a prefix length of 10/xv bases, whereas Fulcrum removed iv.2/4.1 % of duplicates. Equally for CD-Hit-DUP, it removed 3.three/three.0 % of duplicates with up to one/iii mismatches.
As for calculating time, the performance of GPU-DupRemoval to remove identical reads are similar with those obtained by CD-Hitting-DUP and FastUniq, whereas it resulted to be vii.0x faster than Fulcrum. Every bit for most-identical duplicates GPU-DupRemoval resulted be up to one.6x/two.3x faster than CD-Hit-DUP, and up to 12.8x/17.5x faster than Fulcrum to remove duplicates with upward to ane/3 mismatches.
As for retention consumption, a behavior like to the i observed on the SRR921897 library has been observed. In particular, Fulcrum outperforms all remaining tools, requiring in the worst case 1.4 GB of retentiveness, whereas CD-Hit-DUP required 26.9/35.two/37.vii GB of memory to remove duplicates with upward to 0/ane/three mismatches. Equally for GPU-DupRemoval, it required half-dozen.6 GB of memory to remove identical duplicates, and eight.2/6.9 GB to remove nearly-identical ones by clustering the reads with a prefix of 10/15 bases.
Conclusions
In this work we presented GPU-DupRemoval, a tool aimed at removing identical and nearly-identical duplicates from sequencing libraries generated with Illumina platforms. GPU-DupRemoval implements an alignment-gratis strategy and exploits the computational power of modern GPUs to remove duplicates from both single- and paired-stop libraries.
Experimental results show that GPU-DupRemoval is very effective at removing duplicate reads, equally information technology outperforms almost all analyzed tools. In terms of ability to identify and remove duplicates, its functioning are comparable with that of Fulcrum. Withal, it resulted to be very faster than Fulcrum, specially at removing duplicates from paired-cease reads.
The current implementation of GPU-DupRemoval overcomes almost all limitations highlighted with respect to its offset implementation. Currently, the constraint on maximum size of the sequencing library still holds. As highlighted in the previous work, sorting requires all prefixes to be loaded into the memory of the GPU device. Therefore, the maximum size of the library that can exist analyzed depends on the memory of the GPU card used.
Availability and requirements
Project proper noun: GPU-DupRemoval Project home page: http://www.itb.cnr.it/web/bioinformatics/gpu-dupremoval Operating arrangement(southward): LinuxProgramming language: CUDA COther requirements: NVIDIA GPU card with compute capability ≥ 3.5License: free for academic useAny restrictions to utilise past not-academics: license needed
Abbreviations
- CUDA:
-
Compute unified device architecture
- GPU:
-
Graphics processing units
- NGS:
-
Side by side generation sequencing
References
-
Gomez-Alvarez 5, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from circuitous microbial communities. ISME J. 2009; 3(11):1314–vii.
-
DePristo MA, Banks East, Poplin R, Garimella KV, Maguire JR, Hartl C, et al.A framework for variation discovery and genotyping using next-generation Dna sequencing information. Nat Genet. 2011; 43(5):491–8.
-
Li R, Li Y, Fang X, Yang H, Wang J, Kristiansen Thousand, et al.SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009; nineteen(six):1124–32.
-
Magi A, Tattini L, Pippucci T, Torricelli F, Benelli M. Read count approach for Deoxyribonucleic acid copy number variants detection. Bioinformatics. 2012; 28(4):470–8.
-
Zhou X, Rokas A. Prevention, diagnosis and treatment of high-throughput sequencing data pathologies. Mol Ecol. 2014; 23(seven):1679–700.
-
Picard MarkDuplicates. Bachelor from http://broadinstitute.github.io/picard/.
-
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al.The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25(sixteen):2078–nine.
-
Pireddu L, Leo S, Zanetti K. SEAL: a distributed short read mapping and duplicate removal tool. Bioinformatics. 2011; 27(15):2159–sixty.
-
Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009; 25(14):1754–60.
-
Fastx-Toolkit Collapser. Available from http://hannonlab.cshl.edu/fastx_toolkit/.
-
Xu H, Luo 10, Qian J, Pang X, Vocal J, Qian One thousand, et al.FastUniq: a fast de novo duplicates removal tool for paired short reads. PLoS ONE. 2012; vii(12):e52249.
-
Burriesci MS, Lehnert EM, Pringle JR. Fulcrum: condensing redundant reads from high-throughput sequencing studies. Bioinformatics. 2012; 28(10):1324–27.
-
Li W, Godzik A. Cd-hit: a fast program for clustering and comparing big sets of protein or nucleotide sequences. Bioinformatics. 2006; 22(13):1658–9.
-
Fu L, Niu B, Zhu Z, Wu Due south, Li W. CD-Hitting: accelerated for clustering the side by side-generation sequencing data. Bioinformatics. 2012; 28(23):3150–two.
-
Li Westward, Fu L, Niu B, Wu S, Wooley J. Ultrafast clustering algorithms for metagenomic sequence assay. Cursory Bioinform. 2012; 13(6):656–68.
-
Manconi A, Manca Eastward, Moscatelli M, Gnocchi M, Orro A, Armano G, et al. Thousand-CNV: a GPU-based tool for preparing data to detect CNVs with read-depth methods. Front Bioeng Biotechnol. 2015; 3(28):28.
-
Manavski SA, Valle G. CUDA compatible GPU cards as efficient hardware accelerators for Smith-Waterman sequence alignment. BMC Bioinforma. 2008; 9(Suppl two):S10.
-
Luo R, Wong T, Zhu J, Liu CM, Zhu X, Wu East, et al.SOAP3-dp: fast, accurate and sensitive GPU-based curt read aligner. PLoS ONE. 2013; viii(5):e65632.
-
Zhao K, Chu X. M-BLASTN: accelerating nucleotide alignment by graphics processors. Bioinformatics. 2014; 30(10):1384–91.
-
Klus P, Lam Southward, Lyberg D, Cheung MS, Pullan G, McFarlane I, et al.BarraCUDA-a fast short read sequence aligner using graphics processing units. BMC Res Notes. 2012; 5(one):27.
-
Yung LS, Yang C, Wan X, Yu W. GBOOST: a GPU-based tool for detecting gene–cistron interactions in genome–wide case command studies. Bioinformatics. 2011; 27(ix):1309–x.
-
Nvidia-CUDA. Compute unified device architecture programming guide. http://docs.nvidia.com/cuda/index.html.
-
The opencl specification. 2015. https://world wide web.khronos.org/registry/cl/specs/opencl-2.1.pdf.
-
Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008; 36(16):e105.
-
Manca E, Manconi A, Orro A, Armano G, Milanesi Fifty. CUDA-quicksort: an improved GPU-based implementation of quicksort. Concurrency Comput Pract Experience. 2016; 28(one):21–43.
-
Hoberock J, Bell N. Thrust: A parallel template library; 2010. http://thrust.googlecode.com.
-
Sherman Simulator. http://www.bioinformatics.babraham.ac.uk/projects/sherman/.
Acknowledgements
Not applicable.
Declarations
This article has been published as part of BMC Bioinformatics Vol 17 Suppl 12 2016: Italian Gild of Bioinformatics ($.25): Annual Meeting 2015. The full contents of the supplement are available online at http://bmcbioinformatics.biomedcentral.com/articles/supplements/volume-17-supplement-12.
Funding
The work has been supported past the Italian Ministry building of Education and Research through the Flagship InterOmics (PB05) project, by the Lombardy Region through the FRRB grant, and the European MIMOmics (305280) and ELIXIR (https://www.elixir-europe.org) projects. Publication costs have been funded by the Flagship InterOmics project.
Availability of data and material
Not applicable.
Authors' contributions
Conceived the tool: AM. Conceived and designed the experiments: AM, LM. Performed the experiments: AM, MG, GA. Analyzed the information: AM, AO, GA, MM, MG, LM. Wrote the manuscript: AM. Revised the manuscript: AM, GA, LM. Wrote the programme: AM. Generated the synthetic data: AM. Coordinated the project: LM. All authors read and approved the terminal manuscript.
Authors' information
Not applicable.
Competing interests
The authors declare that they accept no competing interests.
Consent for publication
Not applicable.
Ideals approval and consent to participate
Not applicable.
Author data
Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Eatables Attribution iv.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you requite appropriate credit to the original author(s) and the source, provide a link to the Artistic Eatables license, and betoken if changes were made. The Creative Commons Public Domain Dedication waiver(http://creativecommons.org/publicdomain/zero/1.0/) applies to the information fabricated available in this article, unless otherwise stated.
Reprints and Permissions
About this article
Cite this article
Manconi, A., Moscatelli, M., Armano, G. et al. Removing duplicate reads using graphics processing units. BMC Bioinformatics 17, 346 (2016). https://doi.org/10.1186/s12859-016-1192-5
-
Published:
-
DOI : https://doi.org/10.1186/s12859-016-1192-v
Keywords
- Adjacent generation sequencing
- Duplicate reads
- Graphics processing units
- CUDA
How To Filter Duplicate Contig,
Source: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-016-1192-5
Posted by: jorgensenbouselt.blogspot.com


0 Response to "How To Filter Duplicate Contig"
Post a Comment